PCR a strong solution to the experimental pollution! Pract
时间:2018-05-02 来源:EASTWIN浏览次数:53
1 find the source of pollution
If contamination occurs carelessly, we should proceed from the following items and analyze them one by one to exclude pollution.
1. Establishment of Yin Yang control: it is conducive to monitoring the pollution of each component of the reaction system. When the positive control is selected, the weak and reproducible samples should be selected, and a large number of unnecessary amplification sequences can be produced because of strong positive control. Instead, it may become a potential source of pollution. If the recombinant plasmid containing the target sequence is used as a control, the target sequences within the 100 copies are sufficient to produce positive amplification. The selection of negative controls should also be prudent, because PCR is very sensitive and can detect very trace amounts of target molecules from other methods (Sourthern blot or dot blot) negative specimens. In addition, each amplification should include the timing of the reagents in the PCR system, that is, all the components needed for the PCR reaction, without the template DNA, which is very beneficial to the residual pollution of the PCR products in the monitoring reagents. If the reagent in the amplification result is positive, it means that one or several reagents are contaminated. At this time, a batch of new reagents should be replaced with a different reacting tube, each of which contains a test reagent and should be treated immediately after the contamination reagents are detected.
2 environmental pollution
In addition to the possibility of removing the reagent pollution, if the reagents have been found to be contaminated soon after the reagents are replaced, it is possible for the environmental pollution to be considered if the precautions are relatively tight. The main sources of pollution in environmental pollution are as follows:
1. template extraction vacuum drying device;
2. gel electrophoretic collector;
3. electrophoretic device;
4. ultraviolet analyzer;
5. cutting knife or surgical blade;
6. centrifuge;
7. refrigerator door handle, freezing rack, door handle or experimental table.
Wiping experiments can be used to find suspicious sources.
1) wipe the suspicious sources of pollution with sterile cotton swabs that are soaked in sterile water.
2) 0.1ml deionized water soak;
3) take 5ml to do PCR experiment.
4) electrophoretic test results.
8. aerosols. If the above tracking experiment can not find the exact source of pollution, the pollution may be caused by the aerosol of the PCR product in the air. The experimental site should be replaced at this time. If the conditions are not allowed, the new primers are redesigned (no correlation with the original primers).
3 pollution treatment
Environmental pollution
1. dilute acid treatment: scrubbing or soaking the suspected utensils with 1mol/L hydrochloric acid to deactivate the residual DNA; 2. ultraviolet radiation (UV) method: UV wavelength (nm) generally selects 254/300nm and irradiate 30min. It is important to note that the selection of UV to eliminate the contamination of residual PCR products should consider the length of the PCR products and the distribution of bases in the sequence of the products. The UV irradiation is only effective for the long fragments above 500bp, and the effect of the short fragment is not significant. When UV is irradiated, the pyrimidine base in the PCR product forms a two polymer, and these two polymers can terminate the extension, but not all pyrimidine in the DNA chain can form a two polymer, and the UV irradiation can also break the two polymer. The degree of formation of the two dimers depends on the UV wavelength, the type of the pyrimidine two polymer and the sequence of the neighboring nucleotides with the two polymer site. On the long DNA chain irradiated, the number of two polymer defects is less than that of 0.065/ base. The illumination damage of other non two polymer (such as the CyA pyrimidine complex, thymine ethylene glycol, the interchain crosslink between DNA and the DNA fracture) can terminate the extension of the Taq DNA polymerase. The number of these loci is equal to that of the two polymer. If these loci (0.13/ bases) are randomly distributed on the DNA molecule, there will be 32 damage sites on the DNA chain of a 500bp fragment, then each of the 105 molecules will have at least one damage. On the contrary, if the fragment of 100bp has only 6 lesions on each strand, there will be no damage to many molecules in the 105 copy molecules. This is why UV irradiation has a limited fragment length.
Reaction liquid pollution
One of the following methods can be dealt with:
1. DNase I method: PCR mixture (without template and Taq polymerase) was added to 0.5U DNase I, and then heated and inactivated after reaction at room temperature. Then the template and Taq polymerase were added to normal PCR amplification. The advantage of this method is that there is no need to know the sequence of contaminated DNA.
2. endonuclease method: choose the endonucleases (such as Msp I and Taq I) to identify 4 bases, and select several kinds of enzymes at the same time, in order to take one kind of enzyme to identify the defects of specific sequence, and to heat inactivation after 1h at room temperature for PCR; 3. UV irradiation method: UV irradiation without template and Taq polymerase, attention to ultraviolet radiation, attention The items and methods are the same as the above UV irradiation method.
4. g ray radiation method: 1.5kGy radiation can completely destroy the 0.1ng genome DNA, 2 kGy can destroy 104 copies of the plasmid molecules, 4 kGy still does not affect the PCR, but higher than that limit will reduce the efficiency of PCR amplification. Primers can be irradiated without affecting PCR, and G rays destroy DNA by producing free radicals through ionization of water.
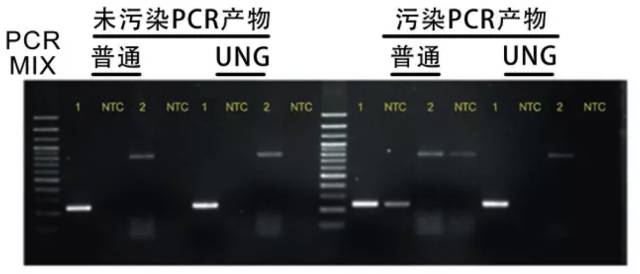
Uridine glucosidase (UNG) method
The effect of UV irradiation on the fragments below 500bp is not good, and the PCR amplification fragment in clinical detection is usually about 300bp. Therefore, the preventive effect of UNG is becoming more and more important and affirmative.
1. principle: in PCR products or primers, dU is used instead of dT. This dU PCR product is incubated with UNG, because UDG can cleave the N- glycosyl bond between the uracil base and the glycosylphosphate skeleton, which can remove the dU and prevent the extension of the TaqDNA polymerase, thus losing the ability to be amplified. UNG has no effect on a template that does not contain dU. UNG can remove uracil from single or double stranded DNA, but has no effect on uracil and single uracil in RNA.
2. dUTP method: replace dTTP with dUTP, so that a large amount of dU can be mixed into the product. Before PCR amplification, the residual contamination of PCR products can be eliminated by treating PCR mixture with UNG. Because UNG can be inactivated in the PCR cycle, it will not affect the new PCR products containing dU.
3. dU primers: when primers were synthesized, dU was used to replace dT, so that only 5 of the PCR products had dU. After UNG treatment, primers lost their binding sites and could not be amplified. The amplification of long fragments (above 1-2kb) is less efficient than that of dUTP, while the dU method can overcome this shortcoming. This method is more efficient than dTTP. The dU primers were best designed for the design of dU at the 3 or near ends. This method can only be used for the treatment of reagents other than primers.
4. advantages: it can remove pollution from any source; UNG treatment can be carried out in the same reaction tube with PCR, and the pollution source can be eliminated completely because of the presence of a large number of dU in the amplification products.
5. it is important to note that DNA doped with dUTP does not affect any operation of the product. When the PCR product is cloned, the UNG- (UNG defect) Escherichia coli receptor should be transformed, otherwise the transformation product will be digested by the receptor bacteria UNG.
Solid phase capture
The principles used to remove contaminated nucleic acids and impurities in specimens are as follows:
1) a single stranded RNA probe with a biotin labelled DNA was hybridized with the nucleic acid to be expanded. The hybridization region was a non amplification region.
2) using the solid phase vector coated with streptavidin to capture hybridization nucleic acids with biotin probes, and remove contaminated amplified products and impurities by rinsing.
3) after the target molecules were eluted, the non RNA probe hybridization region was amplified by specific primers. Second) step PCR can be used to determine whether the specimen is contaminated by amplified products or recombinant plasmids.
RS-PCR method (RNA-specific PCR)
Also known as chain specific PCR, it mainly refers to the specific PCR method used for RNA templates. This method can significantly reduce the false positive without affecting the sensitivity of PCR. The key to the design is to design primers, and the 3 nucleotides of the reverse transcriptase (A region) have 20 nucleotides specific interspecific sequences, and 20 nucleotides (C regions) at the 5 end of the primer are added to the modified base. After mRNA reverse transcriptase, cDNA was separated from superfluous primers by overspeed centrifugation, and then second primers (C) were used to synthesize second chain cDNA with the first chain cDNA as a template. The B region and primer C of the reverse transcriptase primers were amplified and amplified in the subsequent PCR cycle, while the contaminated DNA or plasmid DNA would not be amplified.
Antifouling primer
The primers were cloned by virus DNA, such as plasmid sites. In this region, only intact virus is present. In the recombinant plasmid, the region is divided into two regions and the clone site is obtained. If the recombinant plasmid pollutes the specimen, no bands can be amplified. Even if the amplification band is present, the size is different from what is expected. Only the original virus DNA can be amplified by primers, so the specimen is positive as long as the expected size of the amplified band appears, and this method is used for the ring target molecule series.
(statement: content from the Internet, for sharing purposes only, copyright belongs to the original author. For copyright issues, please contact Xiaobian, delete or delete.
- Previous:Nothing
- Next:Common problems and answers to PCR instrument To learn more Technical data>>>

 English
English 简体中文
简体中文




